 Apichart Linhananta
Apichart Linhananta  Apichart Linhananta
Apichart Linhananta Physics Department, Lakehead University
955 Oliver Road, Thunder Bay, Ontario, Canada
P7B 5E1
Office: CB-4025, Tel:(807) 343 8016,
Fax: (807) 346 7853
apichart.linhananta at lakeheadu.ca
Associate Professor, Lakehead University (2007-present)
Assistant Professor, Lakehead University (2002-2007)
Postdoctoral Associate, SUNY at Buffalo (2000-2002)
Postdoctoral Fellow, Deakin University, Australia (1998-2000)
I. Theoretical Study of Proteins and Biological Systems
Since Levinthal's initial inquiry (1968), protein folding has continued to fascinate biologists, chemists and physicists. Simple theoretical models have illuminate generic aspects of folding, but many properties remain mysterious. We've developed an all-atom model, in which the atoms of a protein are reprsented by a chain of bonded hard spheres. The hard spheres interact by discountinuous square-well and hard-spere interactions, as well as by discontinuous dihedral interactions. Hence, the model is structurally realistic, but is still efficient enough to fold small proteins on a personal computer. The model has successfully been applied to examine the cooperativity of folding [6] as well as the folding mechanism of small helical proteins [1-3] and a beta-hairpin motif [4]. Our work on the helical protein has provided unexpected insights to domain swapping [3], a poorly understood form of protein aggregation and a possible cause of protein folding diseases. I plan to further modify the model and apply it to study the folding of a single protein as well as the interaction between proteins. However, the scope of discontinuous models is limitless. It is highly efficient and is applicable to very complex systems. In addition, it is very flexible and can be easily modify to study other systems of biomolecules as well as complex fluids. Future research may involve study of bio-membranes, membrane proteins or protein recognitions.
The Folding and Aggregation of the Ultra-Fast-Folding
Mini-Protein Trp-Cage
In organisms, newly expressed proteins begin in the unfolded random-coil state.
The ability of proteins to rapidly fold from the randm-coil
state to their unique native structures
is crucial to the health of all living beings.
Most proteins fold to the native states within a few milliseconds to
a few seconds.
The Trp-Cage is one of the fast-folding protein, and can fold to its native
state within a few mircoseconds.
The Trp-Cage Protein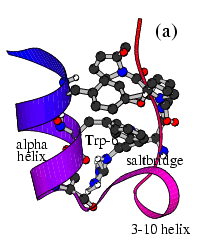
|
Unfolded Random Coil State
|
Folded Native State
|
Fast-Folding Pathway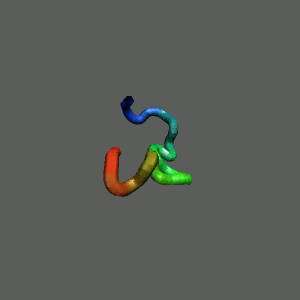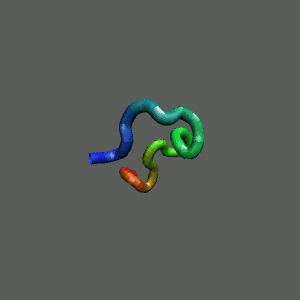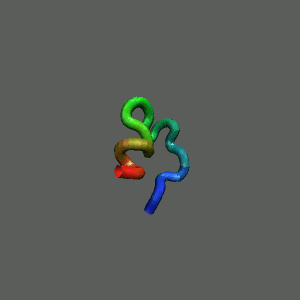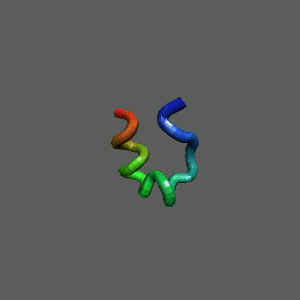 Helix (red segment) forms rapidly, follow by collapse
Helix (red segment) forms rapidly, follow by collapse |
Slow-folding pathway
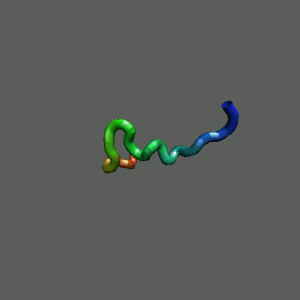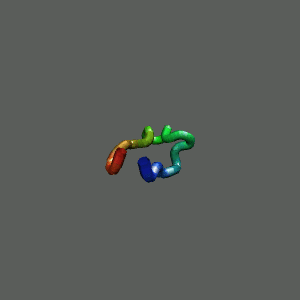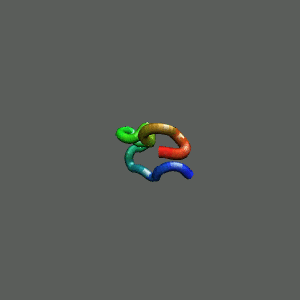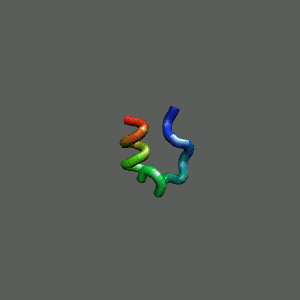 Rapid collapse, follow by the
slow formation of helix Rapid collapse, follow by the
slow formation of helix
|
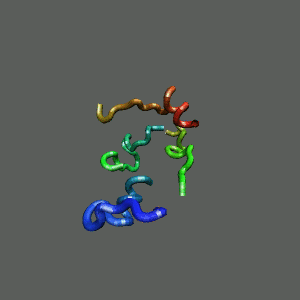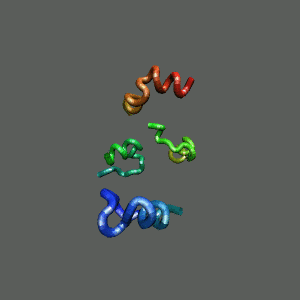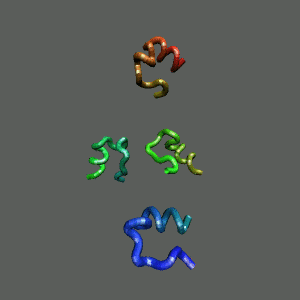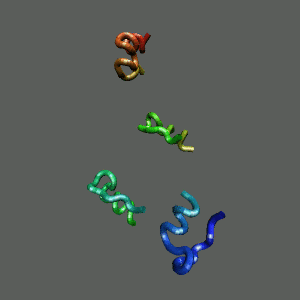
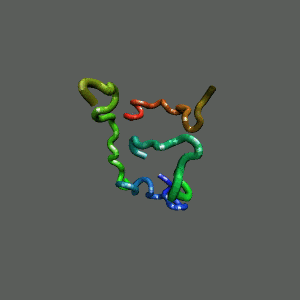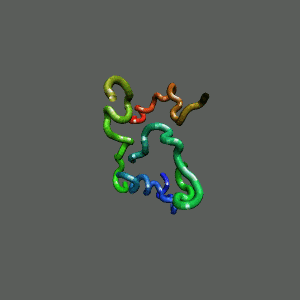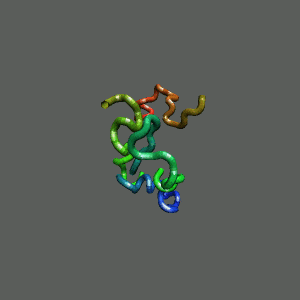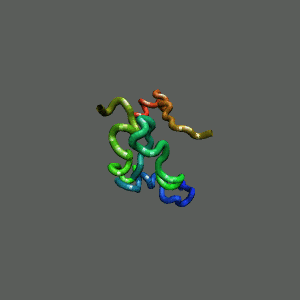
II. Free-Energy Functional Theory of Two-Component Lipid Systems
I am working with Mr. Ian MacKay (former M.Sc. student) on this project. This section is UNDER CONSTRUCTION
III. Collisional Energy Transfer (CET) in Systems of Highly-Excited Hydrocarbons Gas
Quasiclassical trajectory simulations have been performed on highly excited hydrocarbons in rare gas bath. Methods employed include commercial computer codes (VENUS) [6,7], hard-sphere models [4,6], and quantum ab-initio methods. Such simulations provide insights on how hydrocarbon fuels obtain the energy to intitiate combustion reactions. Our works are the first to examine the role of torsional rotors in CET. We found that energy exchange between colliding molecules occur, mainly, via the torsional modes. The "heating up" of the torsional rotors lowers the energy barrier to dissociation reactions, increasing the production of highly reactive radical species that are precusors to combustions.